5th semester

DC1 ALESSANDRO CUZZERI APRIL 25 - September 25
Month 25 (April 25) : Submitted draft of deliverable “An estimation of the microbial biomass and activity originating from the atmosphere and deposited on glacier surfaces”. Prepared submissions for EGU and IMC conferences. Contributed substantially with expertise on bioinformatics to projects referring to biodiversity in snow and ice of alpine ecosystems.
May 2025: EGU attendance with pico presentation. Co-author for talk: Birgit Sattler, Klemens Weisleitner, Patrick Schwenter, and Alessandro Cuzzeri: „Rethinking Glacier Insulation in the Alpine Space: Microplastic Concerns and Sustainable Materials” First part of the DC secondment (Roskilde, Denmark): DNA/RNA extraction of samples from Antarctica 2024 expedition.
June 2025: Completion of lab work in Denmark. Termination and first processing of samples from a second joint project with DC4/Jacobsen CS. Contribution with expertise on bioinformatics for alpine study for microplastics in snow and ice for the Chamber of Commerce.
July 2025 : Sequencing of samples from Antarctica and joint project. Initial data parsing for pesticide mineralization curves analysis. Drafting of own paper (Intra-seasonal trends of cryoconite bacterial communities on an Alpine Glacier).
Month 29 (August 25): Drafting of own paper (Intra-seasonal trends of cryoconite bacterial communities on an Alpine Glacier).
Month 30 (September 2025) International Mountain conference attendance. Oral presentation of pesticides mineralization experiment during a focus session on pollutants in the alpine space. Co-author for talk: Birgit Sattler, Patrick Schwenter, Alessandro Cuzzeri, Monika Summerer, Klemens Weisleitner, Tabea Grube: Slowing Glacial Melt with Sustainable Cellulose Textiles Preparation of the upcoming second part of the DC planned secondment at IGE Grenoble for statistical analyses of Antarctica data.

DC6 KLARA KOHLER May 25 - octcober 25
Month 25: In May, I worked on two different experiments in the lab: I continued working on the timeseries of incubated
sediment experiment, completing the fifth time point. This involved measuring dissolved nutrients in pore water and
supernatant, as well as preparing the sediments for later nutrient extractions. The second experiment involved
crushing different types of sediment and measuring the nutrients released from the sediment before and after
crushing. Also in May, several IceBio students visited our department, where I helped them get started in the lab.
In June, I continued the lab work I had started in May. I also spent some time at ClearWater Sensors in Southampton, UK. Together with Marco Ajmar (DC7), we learned how the ClearWater nutrient sensors are built, how they are deployed, and how sensor maintenance and quality control work. I have also already begun preparations for my next stay at the GFZ in Potsdam to continue the mineralogical work on my sediment samples.
In July, I completed the time series of incubated sediments that I had started in January 2025. As in May, this involved measuring dissolved nutrients and preparing the sediments for nutrient extraction. I also helped Silje Waaler (DC8) preparing her next experiment on nutrient release from glacial flour, which she conducted as part of her secondment in our lab.
In August, I conducted the nutrient extractions on sediment samples from the time series and from the crushing experiment to measure the release of Si, N, P and Fe from the sediment. I also measured major anions at the IC for samples from my completed experiments (incubated sediments, time series and crushing experiments). In August I also visited the GFZ in Potsdam to learn how to perform grain size analysis and specific surface area analysis on my samples.
In September, I began my final fieldwork on the research vessel Polarstern in north-eastern Greenland. Due to the already strong coverage of fast ice, we were unable to go into the fjords and did not reach the mouth of the glaciers to carry out the planned work with the ClearWater sensors. Sampling during the fieldwork was therefore limited to water samples, which were tested for dissolved nutrients, and sediment samples, which were stored for later nutrient extraction analysis back on land.
Month 30: In October, I was still on fieldwork. In addition to my work on the ship, I completed the first draft of my first paper and sent it to my supervisors. I also began to plot the results of my lab work, which I completed in August.

DC7 MARCO AJMAR July 25 - December 25
Month 24 (July 2025): ICP-OES analysis of Iceland 2025 April samples. Prepared for upcoming fieldwork.
Month 25 (August 2025): Fieldwork in Iceland (26/07 – 09/08). Supervised a Master’s student in sampling and sensors deployment. Wrote two deliverables (one about nutrient sensors in glacial environments and the other on nutrient concentrations in glacial meltwaters).
Month 26 (September 2025): Analysed the data collected in August with the nutrient sensors. Organized samples and wrote the method section of my 1st paper.
Month 27 (October 2025): Prepared for the upcoming fieldwork in Iceland, attended the PhD days at the GFZ Potsdam and visited collaborators at the University of Iceland and the Icelandic Met Office.
Month 28 (November 2025): Fieldwork in Iceland (25/10 – 09/11), wrote the 3rd deliverable (about ammonium in situ sensors), attended the GFZ science retreat on writing (2days course).
Month 29 (December 2025) Analysed all Iceland 2025 samples for dissolved macro nutrient species (N, P, Si) and DOC. Supervised a Master’s student for lab analysis and writing. Prepared the PAM 2026 presentation. Attended a time management two days course at the GFZ.
DC4 LARS VAN DIJK August 25 - january 26
Month 25 (August 2025) I established a catalogue of the microbial diversity of the subglacial microbiome as part of Deliverable D2.4.
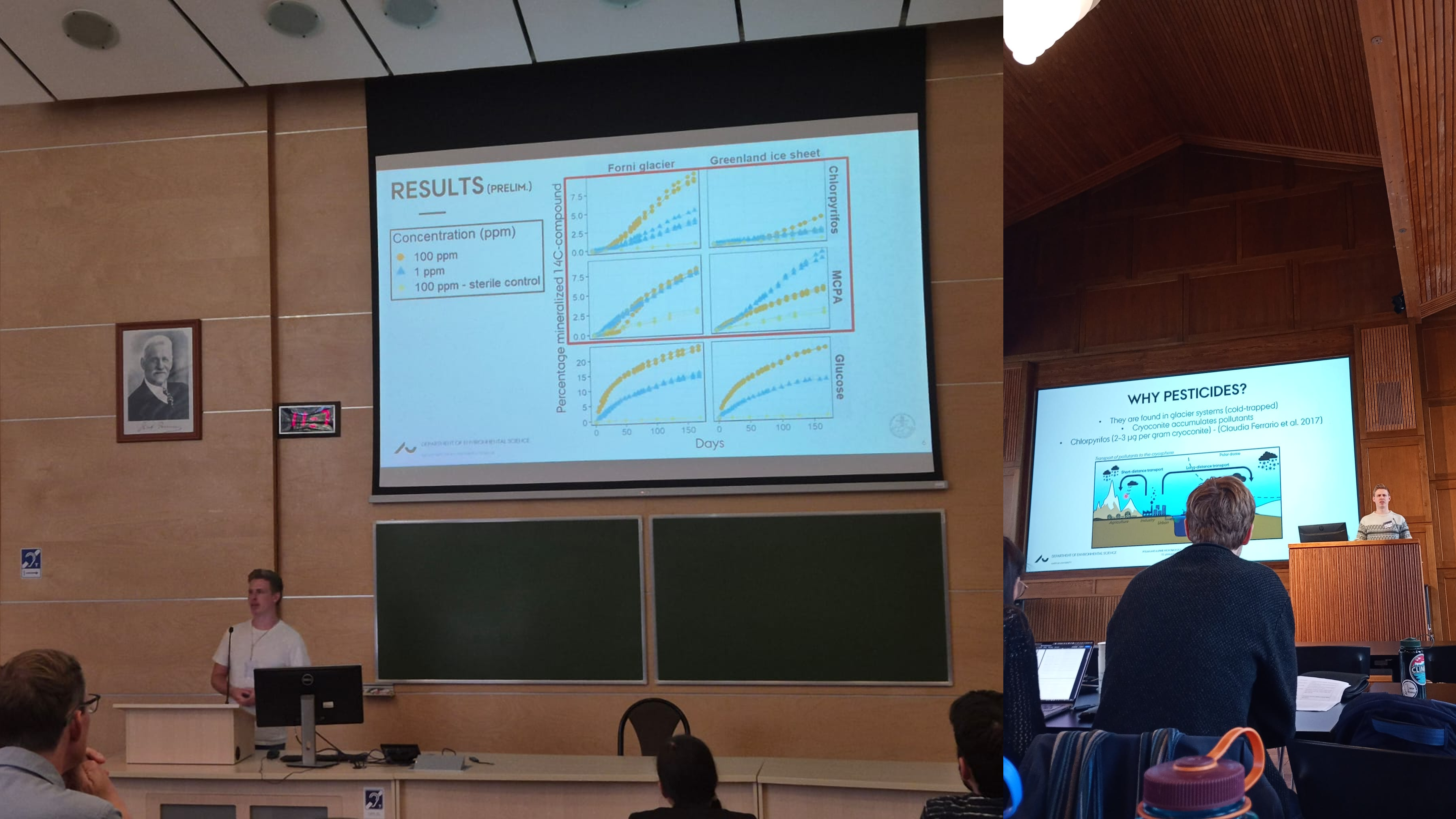
Month 26 September 2025: At the Cryospheric Ecosystems Conference in Poznań, Poland, I delivered an oral presentation, joined useful discussions, and built meaningful connections. I also performed metagenomic sequencing on our pesticide-microcosm samples to complement our total RNA dataset, and plated cryoconite material from these long-term incubations to begin isolating enriched pesticide degraders.
October 2025: I co-organized a workshop on good practices and ethics for using AI in science, focusing on scientific writing and planning. From 16 October to 14 November, I started my secondment with Catherine Larose’s group at the Institut des Géosciences de l’Environnement (IGE) in Grenoble, where I began analyzing the rRNA fraction of our total RNA sequencing data and mapped out workflows for upcoming mRNA, rRNA, and metagenomic analyses.
November 2025: During the secondment at IGE in Grenoble, I continued analyzing the sequencing data and discussed the next steps for the project. I helped initiate and organize the ICEBIO writing camp, to be held the week before the Polar and Alpine Microbiology Conference in Copenhagen. Additionally, this month, candidate pesticide degraders were further purified on agar plates.
December 2025: I continued analyzing total RNA sequencing data obtained from the long-term pesticide incubation microcosm experiment.
Month 30 (January 2026): Due to weather-related disruptions, I attended the ICEBIO writing camp remotely and then participated in the Polar and Alpine Microbiology Conference in Copenhagen on-site, where I gave an oral presentation on the data acquired from the microcosm experiment. Alongside this, I maintained my cyanobacterial and pesticide-degrading cultures and transferred isolates to liquid media for continued growth and eventually Sanger sequencing for identification with the aim to do genome sequencing on unique species.

DC3 ANNIKA MORISCHE August 25 - JAN 26
Month 25 (August 25) started after fieldwork in Kangerlussuaq, from which I brought back 13C-incubated surface ice samples. With these samples I hope to enhance my understanding of how ice algae metabolism works and what metabolites they produce. After safely returning the samples to Denmark, I catalogued and documented the material to prepare it for further MS analyses. At the same time, I was happy to learn that the first batch of similar samples collected in the previous year, which I had extracted and sent frozen to a collaboration partner, arrived successfully and were ready to be analyzed. As a result, the first month involved intensive reading and preparation for starting and advancing my second paper looking into the metabolic range of ice surface autotrophs.
In Month 26, I streamlined my analysis pipeline, spanning raw mass spectrometry data processing through to molecular network analysis. This allowed the integration of molecular information into the statistical analyses of Paper 1, which focuses on the metabolic stress response of surface ice communities to abiotic stress.
Month 27was strongly influenced by research exchange with mass spectrometry experts, leading to new ideas for sample preparation for Paper 3. Based on these discussions, a water-leaching strategy was agreed upon to characterize intracellular metabolites and particulate organic matter from Arctic Ocean winter samples.
Month 28: This led into Month 4, during which the first test samples were selected and prepared. In addition, planning intensified regarding participation organization in and of the Polar and Alpine Microbiology Conference in Copenhagen. Links between this work and my activities within APECs were forged, including the organization of a networking event during the conference.
Month 29: Paper 2 made strong progress. Initial data analysis showed that 13C incorporation was reproducible and exceeded what could be demonstrated using test samples alone. Incorporation of 13C into ice algal pigments could be confirmed, forming the basis for developing a more detailed manuscript focusing on metabolic activity and carbon incorporation in surface ice algae.
Month 30 (January 2026) was dedicated entirely to Paper 1, bringing together all datasets and conducting in-depth statistical analyses to develop an advanced manuscript draft. During this month, the Polar and Alpine Microbiology Conference also took place, where I presented this work and received extensive and valuable feedback, which helped refine the interpretation and strengthen the overall response paper draft.

DC5 ANIRBAN MAJUMDER JULY 25 - Dec 25
Month 25 (July 2025): Most of the time, I was in the lab extracting DNA and RNA from seven fieldwork samples (Svalbard, Greenland, Iceland and Finse). I was preparing for the next slot of metabolite extraction. I was also writing the methods of my second paper.
Month 26 (Aug 2025): This month, I was in the lab performing metabolite extraction from five fieldwork samples (Svalbard, Greenland, Iceland and Finse). I also measured the samples in the mass spectrometry with our collaborator and discussed the further plan for the metabolomics data analysis.
Month 27 (Sept 2025): I was mainly focusing on data analysis and learning the new bioinformatics software required for the analysis of metabolomics and amplicon sequencing.
Month 28 (Oct 2025): I was working on the amplicon sequencing and metabolomics analyses to shape the preliminary story for my future research articles on Svalbard and Greenland microbiomes and metabolites.
Month 29 (Nov 2025): I was in secondment to perform more detailed bioinformatics data analysis (sequencing and mass spectrometry). I also worked on the methodology for my second paper. I submitted an abstract for the Polar and Alpine Microbiology conference (PAM 2026). I also submitted two Icebio deliverables- 1. Deliverable WP1, D1.2, D4 - A catalogue of metabolites in glacier surface habitats (M24, GFZ). 2. D19, D2.6 A catalogue of metabolites in subglacial habitats (M37, GFZ)
Month 30 (Dec 2025): I continued my secondment with bioinformatics data analysis from sequencing and metabolomics. I prepared a first draft of my presentation for the PAM conference to be held in January in Copenhagen.






